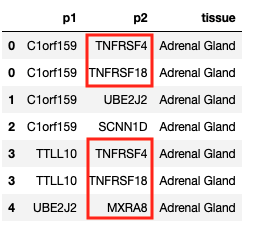
Splitting a row into multiple rows based on substring in a specific column
Here I have a dataframe with 3 columns (p1, p2 and tissue). In column p2 I have 2 genes separated by ';'
1
2
3
4
5
import pandas as pd
pp = pd.read_csv('promoter_promoter.csv', skiprows=2, names=['p1','p2','tissue'])
pp.head()

You can split each of those rows into separate rows by:
1
2
3
pp = pp.assign(p2=pp["p2"].str.split(";")).explode("p2")
pp.head(n=7)
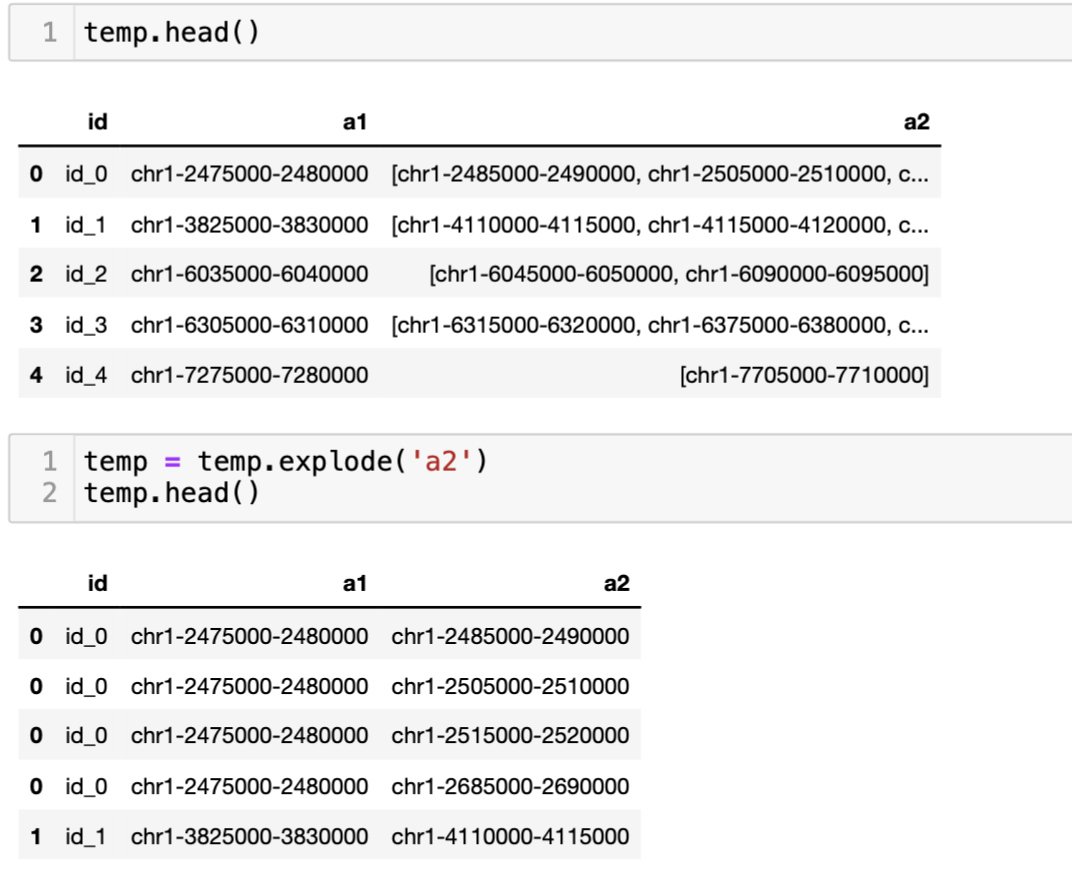
You can just use explode if the values are list. Inn the example below is the interaction data where one regions (a1) is interacting with multiple region (a2) which are presented in a list
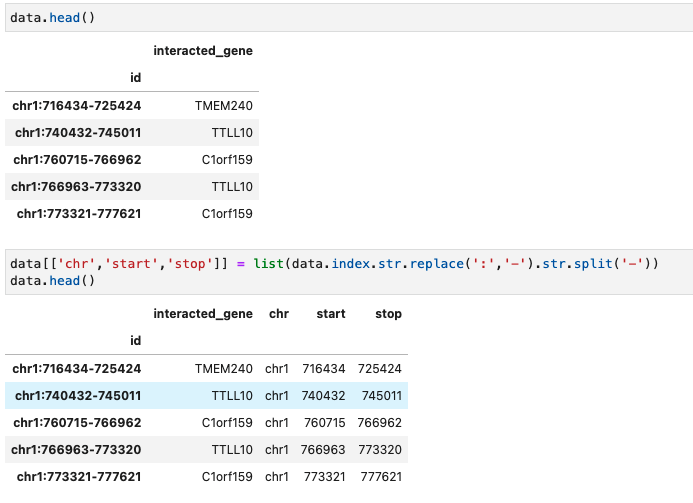
Splitting index into separate columns
Lets say, we have a dataframe of interaction data with genomic region and gene it is interacting with. As shown in the figure below, the genomic coordinates is the index, and we want to split the index into 3 separate columns which can be done by:
data[['chr','start','stop']] = list(data.index.str.replace(':','-').str.split('-'))
Filter out the rows only selecting minimum value based on a specific column
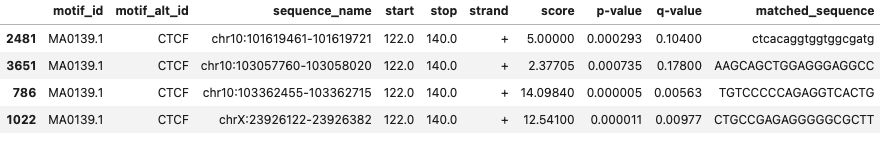
I mainly use this to filter out the output file from program fimo. While scanning for motifs in a given sequnce, there can be multiple region within the sequence where a motif can be found. And I only want to keep those regions with the most significant motifs if there are multiple motifs identified in a given sequence. Here, in this dataframe, chr10:103057760-103058020 and chrX:23926122-23926382 has motifs in two positions and chr10:103362455-103362715 in three.
1
2
3
4
5
import pandas as pd
fimo_df = pd.read_csv('fimo.tsv',sep='\t').iloc[0:-3,:]
fimo_df.head()
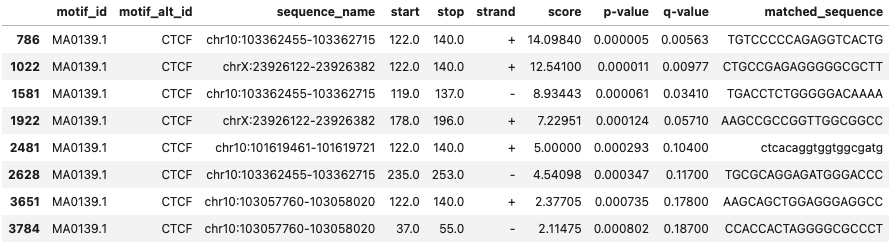
I am using iloc to remove last 3 rows from the fimo output file, which contains program run information. Now, since there are mulitple values for same sequence_name, I will only be keeping the one with lowest p-value
1
2
3
fimo_df = fimo_df.loc[fimo_df.groupby('sequence_name')['p-value'].idxmin()]
fimo_df.head()
Filtering dataframe if substring in a column matches to any elements of a list
Lets assume we have a dataframe as below:
df.head() 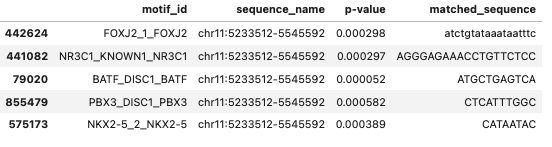
And we want to filter out the dataframe based on our list of transcription factors (TF) as: tf_list = ['BATF','PBX3']
Since, the TF we are interested is a substring in column motif_id, we can’t use df[df['motif_id']].isin(tf_list). So, we can achieve that by:

In the code, above we are also using .upper() function to convert all letters to uppercase if there are any lowercase letters.
Merge multiple dataframes based on a column
Here, there are 4 dataframes, and I want to merge them based on the common column name id.
1
2
3
4
5
6
7
8
import pandas as pd
from functools import reduce
df_list = [df1, df2, df3, df4]
merged_df = reduce(lambda left, right: pd.merge(left, right, on=['id'], how='outer'), df_list)
merged_df = merged_df.dropna()
Compare if two columns are equal in pandas dataframe
1
df['column1'].equals(df['column2'])
This will True or False as output.
